- This event has passed.
Event Navigation
Title: Regulation of Kir Channels and GPCRs by Cholesterol and Anionic Lipids
Host: Rachel Martin
Abstract: KirBac1.1 is an inward-rectifier K+ (Kir) channel. Kir channels are classified by their preference for inward K+ current. KirBac1.1 exists as a tetramer and its structural components include a selectivity filter (or outer gate) which binds K+ions and allows them to pass through the bilayer near the rate of free diffusion, a transmembrane helix bundle exhibiting an “inverted tepee” structure, and a cytoplasmic gating domain or “Kir domain”. This cytoplasmic Kir gating domain is coupled to the activation gate (inner gate) at the base of the inner transmembrane region.
We have discovered how bilayer composition modulates the activity and conformational states of KirBac1.1 using Solid-state NMR (SSNMR) spectroscopy partnered with fluorescence assays and Forster Resonance Energy Transfer (FRET). We identified the activating anionic lipid binding pocket and characterized changes in structure and water accessibility associated with channel gating. Through extensive chemical shift assignments, we further identify two states of the channel under gating conditions. One state is activated and conductive while the second state appears to be shallowly inactivated at the selectivity filter (outer gate). We have also identified a third inactivated state in the absence of anionic lipids. We then describe how cholesterol silences the channel at higher levels. To achieve this, we used SSNMR to measure distances between 13C-KirBac1.1 and 2-13C-cholesterol and discovered two unique cholesterol binding pockets in each bilayer leaflet. In the inner leaflet, near the activation gate, the cholesterol binding site is occupied by a persistent cholesterol dimer. This observation explains how cholesterol efficiently crowds out gating lipids under higher (30%) cholesterol concentrations. These findings are independently confirmed via Molecular Dynamics (MD) simulations of the protein in a cholesterol-rich bilayer. We then solved the structure of the KirBac1.1-cholesterol complex using a customized hybrid CHARMM-22 and CHARRM-36 forcefield deployed within Xplor-NIH.
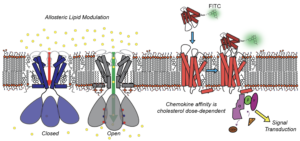
CCR3 is a C-C motif chemokine G protein Couple Receptor (GPCR) expressed by eosinophils and basophils, transducing signals stimulated by the C-C motif chemokine primary messengers 11, 24, and 26. CCR3 is implicated in cancer metastasis, inflammatory conditions, and the cytokine storm. Cholesterol is known to influence GPCR-ligand interactions and receptor multimerization states, introducing a complicated mechanism for homeostatic modulation of GPCR signaling. We present a method using codon harmonization and a Maltose Binding Protein fusion tag for heterologous production of CCR3, obtaining ~3 mg of functional GPCR per liter of minimal media to investigate the effects of cholesterol on CCR3 function in vitro. Affinity for the endogenous ligand CCL11 increases in a dose-dependent manner in both proteoliposomes and lipid nanodiscs using a fluorescence-based functional assay. This is likely coupled to GTPase activity of Gαi3, ostensibly through heightened receptor activation and G protein coupling. This work is validated by both production-quality and course grained MD simulations, revealing key cholesterol binding motifs along the proposed signaling pathway. SSNMR investigation of these functional mechanisms is currently in progress.